The clinical response to the same dose of a drug may vary among individuals. Cytochrome P450 (CYP) 2D6 and 1A2 are enzymes that metabolize many psychotropic medications. Genetic variations in these enzymes may cause changes in their activity and result in differences in effectiveness and adverse effects. Although pharmacogenetic testing is available for CYP2D6 and CYP1A2, interpretation and clinical application of the results may not be straightforward.
Genetic variations in CYP450 enzymes determine enzymatic activity, which can have a large effect on drug levels, efficacy, and toxicity. However, there are many other important factors that clinicians should consider when trying to predict the effects of medications. While clinicians often focus on a patient’s genotype, this only provides information on a chromosomal level, and this information never changes. In contrast, a patient’s phenotype, or status of metabolism, is subject to change throughout the patient’s life.
Many circumstances influence phenotype, including the use of medications that induce or inhibit CYP450 enzymes, environmental factors, and comorbidities. Phenoconversion occurs when these factors result in a phenotype that is different from that predicted by genotype. Because of the possibility of phenoconversion, knowing a patient’s genotype may be of limited value in making clinical decisions. This article provides guidance on interpreting both the genotype and phenotype of CYP2D6 and CYP1A2. Case 1 and Case 2 illustrate these concepts.
CYP2D6
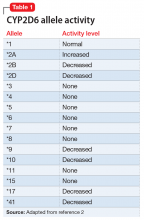
The enzyme activity of CYP2D6 varies among individuals and may include no activity, decreased activity, normal activity, or increased activity. After obtaining the genotype, the activity level of the CYP2D6 alleles may be determined. The frequency with which certain alleles occur varies with ancestry. More than 100 allelic variants and subvariants have been discovered, and new alleles are continuing to be discovered.1Table 12 lists some of the most common CYP2D6 alleles.
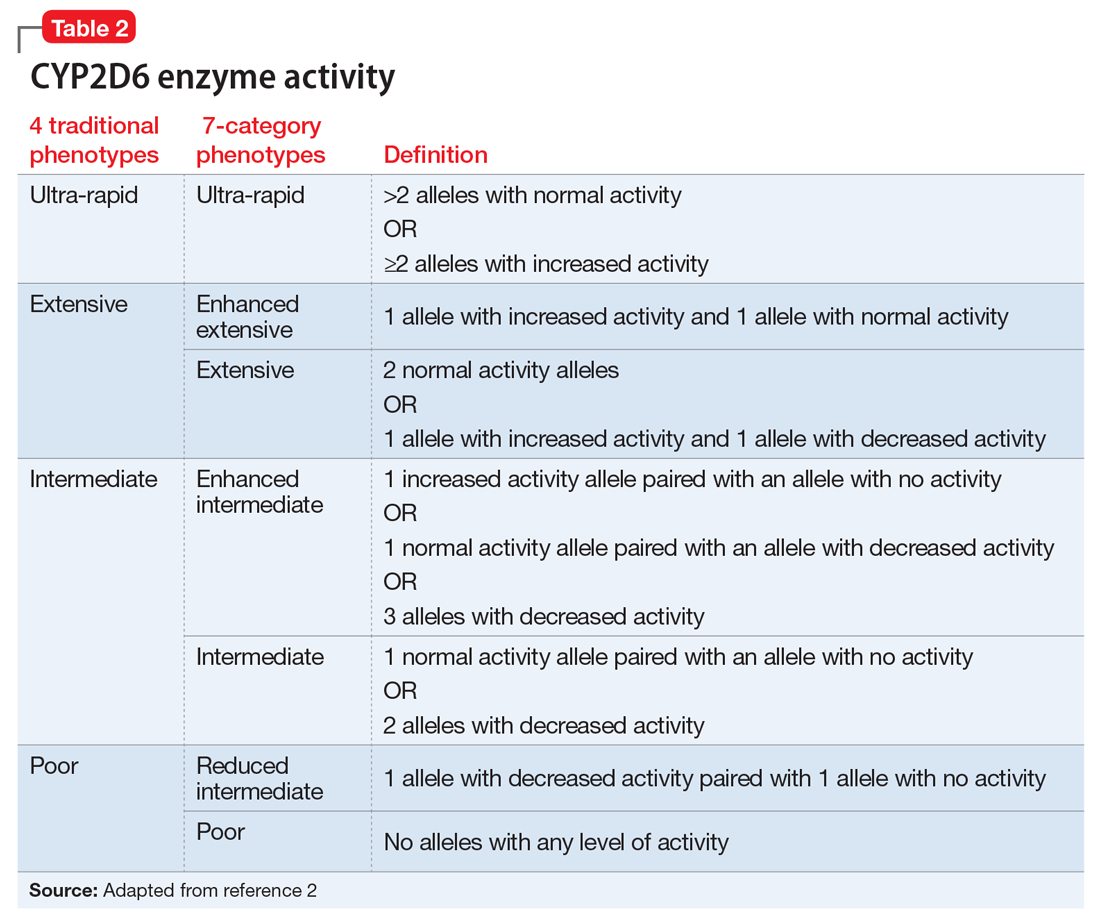
Based on the CYP2D6 enzyme activity determined from the alleles, 4 “traditional” phenotypes can be predicted from the genotype (Table 22). The 7-category phenotypes reported by some laboratory companies provide a more explicit method for reporting phenotypes.
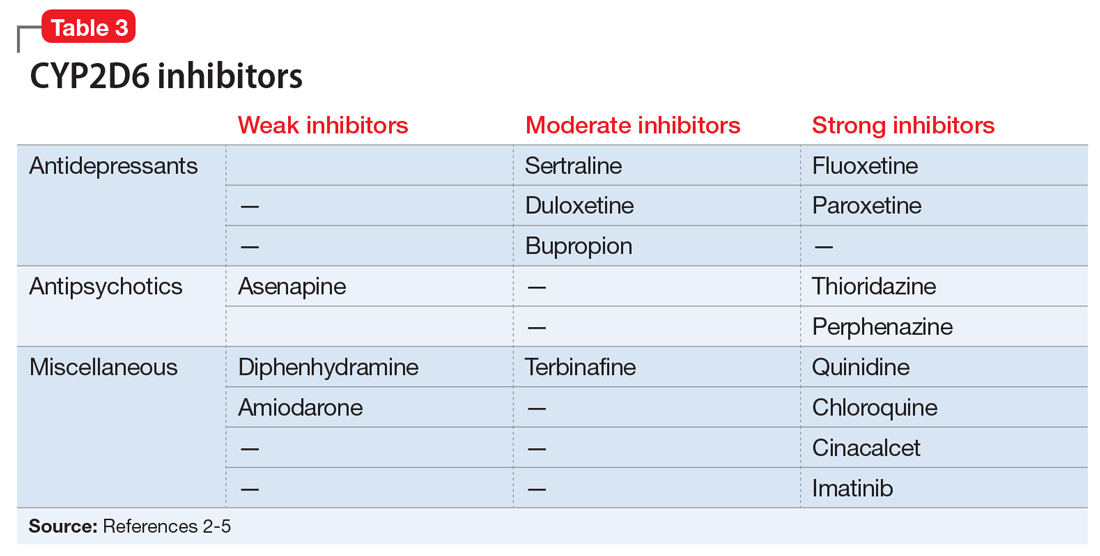
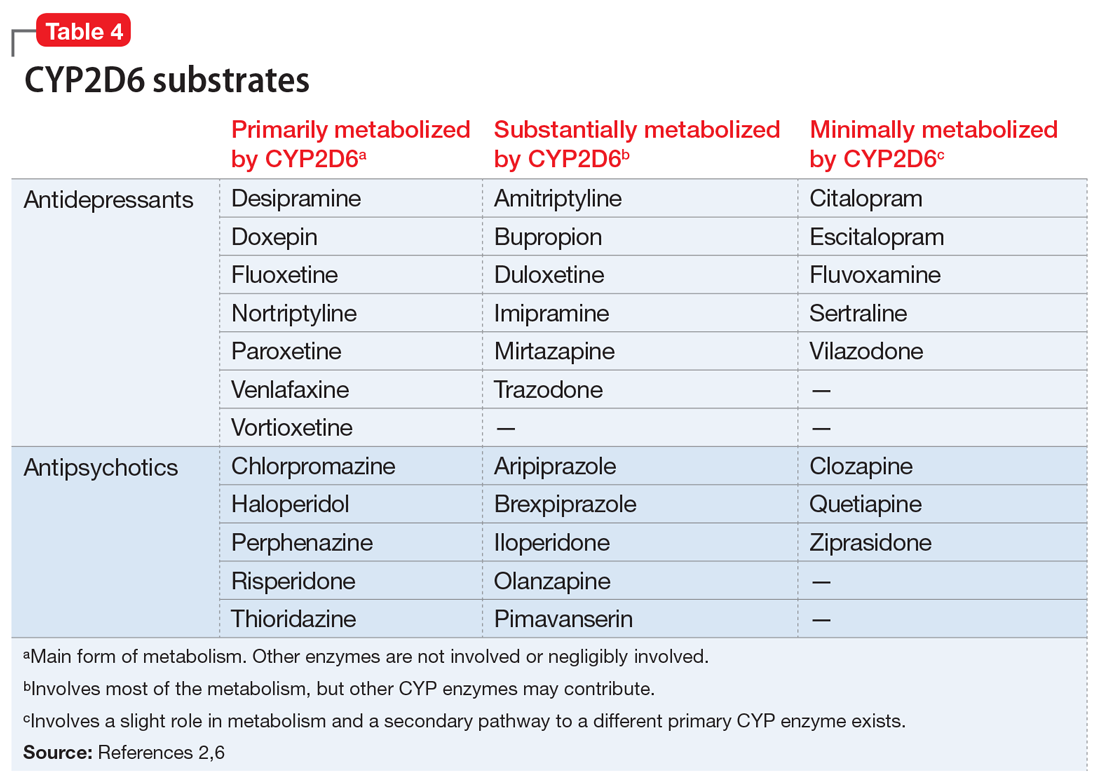
Evidence suggests that, unlike most other CYP450 enzymes, CYP2D6 is not very susceptible to enzyme induction.2 Thus, genetics, rather than drug therapy, accounts for most ultra-rapid CYP2D6 metabolizers. CYP2D6 can be inhibited by the use of medications (Table 32-5) and/or substrates (Table 42,6). Similar to inhibitors, substrates may be saturating high affinity-low capacity enzymes such as CYP2D6, resulting in phenoconversion to poor metabolizers. However, this is unlikely to be the case for substrates of low affinity-high capacity enzymes such as CYP3A4.7 Ultimately, substrates and/or inhibitors of CYP2D6 may result in a phenotype that does not correspond to genotype.
Phenoconversion
Genotyping may not reflect the true prevalence of the CYP2D6 poor metabolizer phenotype when using multiple medications that are substrates and/or inhibitors of CYP2D6.8 In the presence of strong CYP2D6 inhibitors, up to 80% of individuals with a non-poor metabolizer genotype are converted to a poor metabolizer phenotype.8 While the phenotype provides a clearer representation of metabolism status than genotype, this information may not always be available.
Continue to: Determining CYP2D6 phenotype