Systems biology is relatively new. It is an interdisciplinary field that focuses on complex interactions within biological systems using a holistic approach in the pursuit of scientific discovery.
The systems biology approach seeks to integrate biological knowledge to understand how cells and molecules interact with one another. A key component is computational and mathematical modeling. The ever-increasing amount of biological data, and the judgment that this data cannot be understood by simply drawing lines between interacting cells and molecules, explains the demand for a systematic approach.
Prominent examples for biological systems are the immune system and the nervous system, which already have the word ”system” included. Although the idea of system-level understanding is not new, the growing interest in applying the systems approach has been driven by breakthrough advances in molecular biology and bioinformatics.
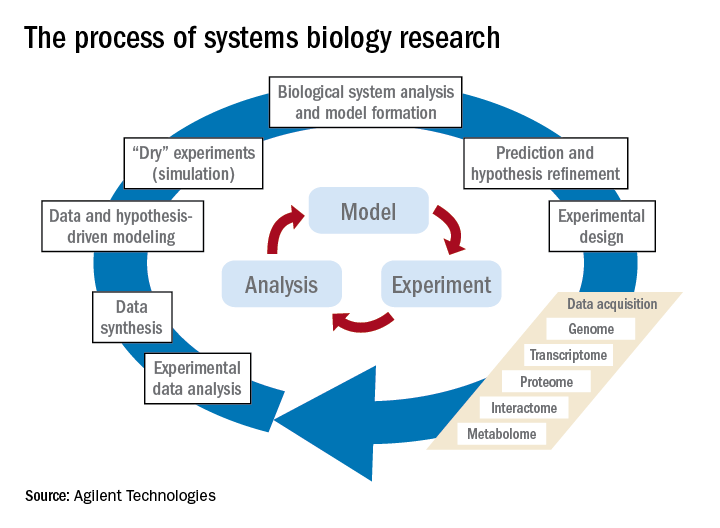
The process of systems biology research involves the steps shown in the figure. In addition to the steps of traditional experiments with hypotheses that lead to data acquisition (now often termed a reductionist approach), the systems approach adds computational and mathematical modeling to integrate the data and biological knowledge to understand how the system acts together within a network. While model building is key in systems biology, it’s also important to harness the ever-expanding amount of biological data in the literature by performing advanced searches; by doing so, that knowledge can be included in the computer modeling of experiments.Over the past 10 years, our group has identified highly significant differences in immune functioning between the 10% of children who frequently develop acute otitis media (i.e., those who are “otitis prone”) and the children who develop AOM infrequently (60% of children) or not at all (30% of children). We also have identified a cohort of about 10% of children who fail to respond to infant vaccinations (low vaccine responders), compared with children who respond with protective immunity and establishment of immune memory. The differences in children who are prone to AOM vs. those who are not and in low vaccine responders vs. normal vaccine responders include differences in cytokine molecules in blood (providing biosignatures), reduced antibodies, immune memory, and aberrant intercellular signaling networks after otopathogen exposure (AOM prone vs. non–AOM prone) and routine pediatric vaccination (low vs. normal vaccine responders).
After searching and compiling more than 30,000 articles in the literature on AOM etiology, pathogenesis, and immune response, as well as more than 30,000 articles on pediatric vaccines and vaccination responses, we have proposed to the National Institutes of Health that the information in this literature and in our body of experimental data be used to assemble a systems network model of the immune circuitry engaged during pathogenesis of AOM and causality of low vaccine responders. This general framework would serve to integrate existing data from previous studies involving children and animal models, mechanistically support network models derived directly from experimental data, and simulate the behavior of these networks to support the gradual refinement of corrective and/or preventative treatments. Keep your fingers crossed.Dr. Pichichero, a specialist in pediatric infectious diseases, is director of the Research Institute at Rochester (N.Y.) General Hospital. He is also a pediatrician at Legacy Pediatrics in Rochester. He has no relevant financial disclosures. Email him at pdnews@frontlinemedcom.com.